Structural Analysis and Modelling Platform
The Structure Analysis & Modelling Platform offers a comprehensive set of spectroscopic, biophysical and computational tools to determine molecular structures and functional mechanisms of the systems under investigation in all 3 Topics. Our main methods include solid-state NMR (Ulrich), liquid-state NMR (Luy), circular dichroism (Bürck), fluorescence techniques (Nienhaus), multiscale molecular dynamics simulations (Elstner), as well as computational modelling (Wenzel).
The biomolecules studied in BIFTM include small compounds and soluble proteins, membrane-bound peptides and transmembrane proteins, dynamic protein-effector complexes and large oligomers. Most applications are focussed on molecules embedded in and related to biomembranes, which constitute the "interface" between every cell and its environment. Membranes have to be overcome as barriers in cell penetration processes and signal transduction events (Topic 1), and they have to be permeabilized for antimicrobial action and biofilm control (Topic 2). Polymers and other amorphous solid materials (Topic 3) are also amenable for study using the techniques available in this Platform such as biodegradable silk fibers or synthetic macromolecules.
This Platform provides not only designated analytical approaches for soluble, soft and solid systems, but also includes unique instrumental facilities with synchrotron radiation, as well as molecular dynamics simulations and computational multiscale modelling methods. These activities are complemented by materials and techniques from the Soft Matter Synthesis Lab (SML) and the Bioinformatics Platform.

NMR spectroscopy: Methods are available for analyzing soluble biomolecules (i.e. effectors and ligands, soluble target proteins < 50 kDa and small membrane proteins in micelles) using liquid-state NMR (at 700, 600 MHz), as well as membrane-bound systems by means of solid-state NMR ( at 600, 500, 300, 200 MHz). Our unique expertise in Karlsruhe lies in oriented samples, which are intrinsically suited for biomembrane preparations, but also are highly advantageous for soluble molecules in weakly aligned matrices. Here, anisotropic nuclear interactions are analyzed as a rich source of information about short-range and long-range molecular geometry.
Synchrotron radiation circular dichroism (SR-CD): we are operating a high-performance vacuum-UV circular

dichroism beamline (UV-CD12) at ANKA as an international user facility, in addition to two conventional desktop CD spectropolarimeters. A set-up for oriented CD (OCD) is available on all instruments thus representing the world´s first SR-OCD beamline.
Circular dichroism with synchrotron radiation as a light source offers a number of advantages compared to conventional benchtop instruments, the most important being the extension of the lower wavelength limit well below 190 nm and the considerably improved signal-to-noise ratio. Access to this extended spectral range allows a far more powerful identification of protein fold motifs. It is possible to discriminate 7-9 distinct motifs compared with the 3-4 types of structures that can be resolved by conventional CD.
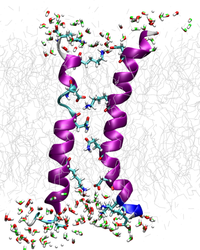 Computational modelling: Quantum mechanical methods are combined with empirical force field methods to treat the functionally relevant site of a protein with QM, while the remaining environment is handled with MM and/or continuum approaches. For membrane systems, coarse-grained models as well as atomistic force field MD simulations are being employed, besides structure-based Go-modelling approaches and continuum methods. We have developed tools for protein structure prediction, protein folding, molecular docking, hydrophobic moment calculations and 3D visualization.
Computational modelling: Quantum mechanical methods are combined with empirical force field methods to treat the functionally relevant site of a protein with QM, while the remaining environment is handled with MM and/or continuum approaches. For membrane systems, coarse-grained models as well as atomistic force field MD simulations are being employed, besides structure-based Go-modelling approaches and continuum methods. We have developed tools for protein structure prediction, protein folding, molecular docking, hydrophobic moment calculations and 3D visualization.
